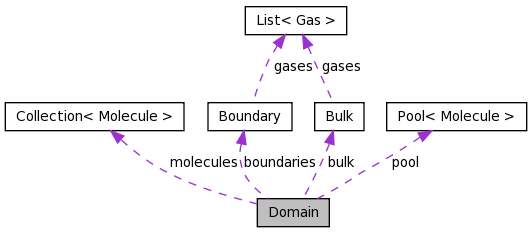
Domain Class Reference
Computational Domain. More...
#include <domain.h>
Public Member Functions | |
| Domain (char *filename) | |
| Domain constructor creates an instance of a domain. | |
| ~Domain () | |
| void | BoundaryType (enum BoundaryTypes b, int idir, int inside) |
| enum BoundaryTypes | BoundaryType (int idir, int inside) |
| void | setMinBound (int i, REAL x) |
| void | setMaxBound (int i, REAL x) |
| REAL | minBound (int i) |
| REAL | maxBound (int i) |
| int | computeBounds (REAL x0[], REAL x1[]) |
| Collection< Molecule > * | Molecules () |
| int | run (int niter) |
| The Domain solver: running niter iterations. | |
| void | step (int iter) |
| int | boundary (Molecule *molecule) |
| void | injection () |
| Inject cross-boundary species:. | |
| void | interaction () |
| void | interaction (int icounter) |
| Interaction | interact (Molecule *a, Molecule *b) |
| void | init () |
| void | load (char *filename) |
| Initializing molecules according to bulk properties (restart=0). | |
| void | save (char *taskname) |
Private Attributes | |
| int * | distribution |
| molecule counts for all species | |
| REAL | bounds [2][DIM] |
| char | boundaryName [maxBoundaryTypes][WORDLENGTH] |
| to be depricated | |
| Bulk | bulk |
| bulk of the domain: initialized only if Run::option.restart=0 | |
| Boundary | boundaries [DIM][2] |
| six boundaries of a hexahedral domain | |
| Collection< Molecule > | molecules |
| Collection of molecules. | |
| Pool< Molecule > * | pool |
| Molecule pointer pool. | |
Detailed Description
Computational Domain.Implements main data types inside a computational domain
Definition at line 83 of file domain.h.
Constructor & Destructor Documentation
| Domain::Domain | ( | char * | filename | ) |
Domain constructor creates an instance of a domain.
get domain specs from XML configfile
read XML input file
Loop over file records
set xterm output mode:
Read the number of molecules:
Read species:
load reactions
Read domain specs:
Read domain bounds
Read initial bulk properties
Read boundary specs
Read boundary reactions:
< probability for the formation of these products as opposed to different products If only one pair of products exists the probability = 1
Definition at line 72 of file domain.cc.
References Species::Reaction::Add(), Boundary::adiabatic, List< Element >::append(), Boundary::area, Geom::area(), AtomicMassUnit, AvogadroNumber, BoltzmannConstant, boundaries, boundary(), boundaryName, bounds, bulk, GridContainer::cellsize, Run::configfile, Species::Specie::Cp(), Collection< Element >::Current(), Species::Gas::density, DIM, distribution, DOCTYPE, elasticBoundary, Species::Reaction::Outcome::Enthalpy(), Species::Reaction::First(), GasConstant, Boundary::gases, Bulk::gases, IO::getCharAttr(), Collection< Element >::goFirst(), Collection< Element >::goNext(), Species::Specie::Id(), init(), Bulk::init(), Boundary::init(), Collection< Element >::init(), insideBoundary, Collection< Element >::isFirst(), Potential::lengthscale(), load(), Species::Specie::Mass(), maxBound(), maxBoundaryTypes, MAXLINLEN, minBound(), molecules, Species::Reaction::Next(), Species::nspecies, Collection< Element >::number(), openBoundary, Run::option, IO::parseFloat(), IO::parseWord(), periodicBoundary, pool, Species::Reaction::Outcome::Probability(), Species::Reaction::Outcome::Product(), GridContainer::put(), Boundary::reactions, Species::reactions, REAL, setMaxBound(), setMinBound(), Species::Specie::Size(), Gui::size, Species::species, Potential::strength(), Boundary::temperature, Bulk::Temperature(), TINY, Boundary::type, VOIDSPECIE, WORDLENGTH, XMAX, Gui::xmax, XMIN, Gui::xmin, and Option::xterm.
| Domain::~Domain | ( | ) |
Member Function Documentation
| int Domain::boundary | ( | Molecule * | molecule | ) |
< do reaction: A + B =
< new molecule A type
< new molecule B type
Definition at line 1076 of file domain.cc.
References boundaries, bounds, Molecule::Coordinates(), Species::Specie::Cp(), DIM, DOF, DOFI, DOFK, Gui::dx, elasticBoundary, Species::Reaction::Outcome::Enthalpy(), Species::Reaction::First(), Collection< Element >::insertAfter(), insideBoundary, Molecule::InternalEnergy(), Molecule::KineticEnergy(), LENGTH, Species::Specie::Mass(), molecules, MULC, Species::Reaction::Next(), openBoundary, periodicBoundary, Species::Reaction::Outcome::Probability(), Species::Reaction::Outcome::Product(), Species::reacted, Boundary::reactions, REAL, RND, Species::Specie::Size(), SMALL, Species::species, Run::time, Species::Reaction::Outcome::Time(), Molecule::Type(), Boundary::type, Molecule::Velocity(), VOIDSPECIE, XMAX, and XMIN.
Referenced by Domain(), injection(), and run().
| enum BoundaryTypes Domain::BoundaryType | ( | int | idir, | |
| int | inside | |||
| ) | [inline] |
| void Domain::BoundaryType | ( | enum BoundaryTypes | b, | |
| int | idir, | |||
| int | inside | |||
| ) | [inline] |
| int Domain::computeBounds | ( | REAL | x0[], | |
| REAL | x1[] | |||
| ) |
Definition at line 829 of file domain.cc.
References Molecule::Coordinates(), Collection< Element >::Current(), DIM, Collection< Element >::goFirst(), Collection< Element >::goNext(), Collection< Element >::isFirst(), LARGE, molecules, and REAL.
| void Domain::init | ( | ) |
create local gas-array
Go over all gases
point to current gas
select current gas-specie
Estimate the average velocity and the number of molecules of each specie to be injected into the bulk
get molecule's mass in atomic units
convert molecule's mass to kg
average velocity in one direction
get density in kg/m^3
get the number of molecules in the bulk
Inject all molecules:
Definition at line 621 of file domain.cc.
References AtomicMassUnit, BoltzmannConstant, bulk, List< Element >::Current(), Species::Gas::density, Bulk::gases, List< Element >::goFirst(), List< Element >::goNext(), Species::Specie::Id(), Bulk::inject(), List< Element >::isFirst(), Species::Specie::Mass(), molecules, List< Element >::number(), Run::option, Species::Gas::specie, Species::species, Molecule::Temperature(), Bulk::Temperature(), and Bulk::Volume().
Referenced by Domain().
| void Domain::injection | ( | ) |
Inject cross-boundary species:.
Inject cross-boundary species. The algorithm uses the density and temperature to caclulate the frequency of injection of molecules of specie, 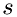 at the boundary. The injection frequency,
at the boundary. The injection frequency, 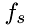 per unit area,
per unit area, 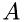 is computed as
is computed as
![\[ f_s = \frac{N}{\Delta\,t} \]](form_5.png)
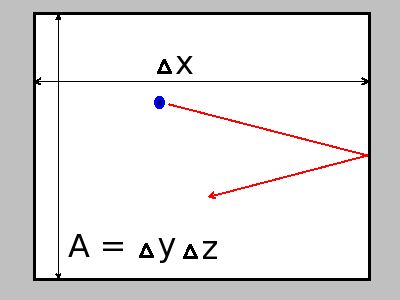
where 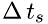 is the time interval at which a molecule hits the boundary area
is the time interval at which a molecule hits the boundary area 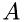 , and
, and 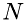 is the number of molecules in a volume with the base
is the number of molecules in a volume with the base 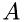 and length
and length 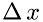 as shown in the figure:
as shown in the figure:
'
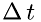 , between the collisions can be related to the component of velocity of the molecule of species
, between the collisions can be related to the component of velocity of the molecule of species 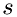 , in direction
, in direction 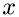 ,
, 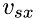 , as follows:
, as follows:
![\[ \Delta\,t = \frac{2\Delta\,x}{v_{sx}} \]](form_19.png)
The number of molecules, 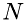 can be related to density,
can be related to density, 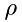 , as:
, as:
![\[ N = \frac{\rho}{\mu}\Delta\,x\,A \]](form_20.png)
where 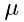 is the mass of one molecule and
is the mass of one molecule and 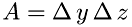 . Thus, the frequency is:
. Thus, the frequency is:
![\[ f_s = \frac{\rho}{\mu}\Delta\,x\,A\frac{v_{sx}}{2\Delta\,x} = \frac{\rho\,v_{sx}}{2\mu}A \]](form_22.png)
get time-step in sec
Go over all 6 boundaries:
select a boundary
get boundry area in m^2
get boundary temperature in K
create local gas-array
Go over all gases
point to current gas
select current gas-specie
Use kinetic theory of gases to estimate collision frequency with the wall and the number of molecules injected. This number can be fractional, so split it into integer and fractional part and inject the fractional part using probability.
get molecule's mass in atomic units
convert molecule's mass to kg
get density in kg/m^3
estimate collision frequency per unit area
calculate the number of collisions with the boundary during the past time step
get the whole integer number of molecules injected
calculate the fractional part of a molecule injected
Inject all whole molecules:
Probabilistically inject the fractional molecules:
Definition at line 1260 of file domain.cc.
References Boundary::area, Geom::area(), AtomicMassUnit, BoltzmannConstant, boundaries, boundary(), Species::Gas::density, DIM, Boundary::gases, List< Element >::goFirst(), Boundary::Inject(), Species::Specie::Mass(), molecules, REAL, RND, Species::Gas::specie, Species::species, step(), Boundary::temperature, and Run::time.
Referenced by run().
| Interaction Domain::interact | ( | Molecule * | a, | |
| Molecule * | b | |||
| ) |
Computes interaction between two molecules with the exchange of energy and momentum according to hard-ball collisions, and the possibility of chemical reactions. Also estimates time to the next collision. Initializing molecules according to bulk properties (restart=0) The velocity of each molecule is set from given temperature according to:
![\[ \frac{3}{2}kT = \frac{mV_{av}^2}{2} \]](form_41.png)
Since the average velocity relates to the average velocity in one direction, x, as:
![\[ V_{av}^2 = 3V_{x,av}^2 \]](form_42.png)
we obtain for 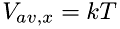
Definition at line 1486 of file domain.cc.
References Species::Reaction::Outcome::ActivationEnergy(), Species::annihalated, Species::collided, Molecule::Coordinates(), Species::Specie::Cp(), DIM, DOF, DOFI, DOFK, Palette::dvi, Gui::dx, Species::Reaction::Outcome::Enthalpy(), Species::Reaction::First(), Species::Specie::Id(), Molecule::InternalEnergy(), Species::Specie::Mass(), Species::missed, Species::Reaction::Next(), Species::nspecies, Species::Reaction::Outcome::Probability(), Species::Reaction::Outcome::Product(), Species::reacted, Species::reactions, REAL, RND, SCLP, Species::Specie::Size(), SMALL, Species::species, Molecule::Type(), Molecule::Velocity(), and VOIDSPECIE.
Referenced by interaction().
| void Domain::interaction | ( | int | icounter | ) |
DDD: dirty fix, should be as below, but did not work
DDD: end dirty fix
DDD: dirty fix, should be as below, but did not work
DDD
DDD: dirty fix: some interactions may go unaccounted
DDD
Definition at line 1383 of file domain.cc.
References Species::annihalated, Container< Element >::Current(), DIM, Gui::dx, Container< Element >::First(), Container< Element >::FirstNext(), Container< Element >::goFirst(), Container< Element >::goNext(), GridContainer::index(), interact(), Container< Element >::isFirst(), Container< Element >::isFirstLast(), Container< Element >::isLast(), Species::missed, GridContainer::ncells, Gui::nodes, Run::nthreads, Container< Element >::number(), REAL, Container< Element >::setFirst(), Container< Element >::setFirstLast(), Run::time, Molecule::Type(), VOIDSPECIE, XMAX, and XMIN.
| void Domain::interaction | ( | ) |
Definition at line 1306 of file domain.cc.
References Species::annihalated, Container< Element >::Current(), DIM, Gui::dx, Container< Element >::First(), Container< Element >::FirstNext(), Container< Element >::goFirst(), Container< Element >::goNext(), GridContainer::index(), interact(), Container< Element >::isFirst(), Container< Element >::isFirstLast(), Container< Element >::isLast(), Species::missed, GridContainer::ncells, Gui::nodes, Container< Element >::number(), REAL, Container< Element >::remove(), Container< Element >::setFirst(), Container< Element >::setFirstLast(), Molecule::Type(), VOIDSPECIE, XMAX, and XMIN.
Referenced by run().
| void Domain::load | ( | char * | filename | ) |
Initializing molecules according to bulk properties (restart=0).
Definition at line 687 of file domain.cc.
References Collection< Element >::append(), Molecule::Coordinate(), Potential::Cutoff(), DIM, Molecule::InternalEnergy(), IO::ioutput, LARGE, Potential::lengthscale(), MAXLINLEN, molecules, Species::nspecies, Collection< Element >::number(), Run::option, REAL, Species::Specie::Size(), Gui::size, Species::species, Run::time, Molecule::Type(), Molecule::Velocity(), and WORDLENGTH.
Referenced by Domain().
| REAL Domain::maxBound | ( | int | i | ) | [inline] |
Definition at line 111 of file domain.h.
References bounds.
Referenced by Domain(), and Gui::readconf().
| REAL Domain::minBound | ( | int | i | ) | [inline] |
Definition at line 110 of file domain.h.
References bounds.
Referenced by Domain(), and Gui::readconf().
| Collection<Molecule>* Domain::Molecules | ( | ) | [inline] |
Definition at line 115 of file domain.h.
References molecules.
Referenced by Gui::display(), and Gui::initdisp().
| int Domain::run | ( | int | niter | ) |
The Domain solver: running niter iterations.
The main loop over niter iterations or till the end of time:
Definition at line 849 of file domain.cc.
References AtomicMassUnit, BoltzmannConstant, boundary(), GridContainer::cellsize, Species::Specie::Cp(), Collection< Element >::Current(), DIM, distribution, DOF, DOFI, ESC, Collection< Element >::goFirst(), Collection< Element >::goNext(), injection(), interaction(), Molecule::InternalEnergy(), IO::ioutput, Collection< Element >::isFirst(), Molecule::KineticEnergy(), LARGE, Collection< Element >::maxCounters(), maxcounters, Collection< Element >::maxnumber(), molecules, Molecule::Move(), GridContainer::nodes, Species::nspecies, Run::nthreads, Collection< Element >::number(), onethird, Run::option, Run::outputname, GridContainer::put(), REAL, Container< Element >::remove(), Collection< Element >::remove0(), save(), SMALL, Species::species, step(), Run::time, twothirds, Molecule::Type(), Molecule::Velocity(), and VOIDSPECIE.
Referenced by Gui::animate(), Gui::keyboard(), main(), and Gui::runmany().
| void Domain::save | ( | char * | taskname | ) |
Definition at line 1935 of file domain.cc.
References Molecule::Coordinate(), Collection< Element >::Current(), Run::elapsed(), Collection< Element >::goFirst(), Collection< Element >::goNext(), Species::Specie::Id(), Molecule::InternalEnergy(), IO::ioutput, Collection< Element >::isFirst(), MAXLINLEN, molecules, Collection< Element >::number(), Run::option, Species::species, Run::time, Molecule::Type(), Molecule::Velocity(), and VOIDSPECIE.
Referenced by Gui::keyboard(), main(), and run().
| void Domain::setMaxBound | ( | int | i, | |
| REAL | x | |||
| ) | [inline] |
Definition at line 109 of file domain.h.
References bounds.
Referenced by Domain(), and Gui::initdisp().
| void Domain::setMinBound | ( | int | i, | |
| REAL | x | |||
| ) | [inline] |
Definition at line 108 of file domain.h.
References bounds.
Referenced by Domain(), and Gui::initdisp().
| void Domain::step | ( | int | iter | ) |
Referenced by injection(), and run().
Member Data Documentation
Boundary Domain::boundaries[DIM][2] [private] |
six boundaries of a hexahedral domain
Definition at line 88 of file domain.h.
Referenced by boundary(), BoundaryType(), Domain(), and injection().
char Domain::boundaryName[maxBoundaryTypes][WORDLENGTH] [private] |
REAL Domain::bounds[2][DIM] [private] |
Definition at line 85 of file domain.h.
Referenced by boundary(), Domain(), maxBound(), minBound(), setMaxBound(), and setMinBound().
Bulk Domain::bulk [private] |
int* Domain::distribution [private] |
Collection<Molecule> Domain::molecules [private] |
Collection of molecules.
Definition at line 89 of file domain.h.
Referenced by boundary(), computeBounds(), Domain(), init(), injection(), load(), Molecules(), run(), and save().
Pool<Molecule>* Domain::pool [private] |
The documentation for this class was generated from the following files:
 1.5.7.1
1.5.7.1